 CAD Software for
DNA Nanostructures with
Axial
Symmetry
CAD Software for
DNA Nanostructures with
Axial
Symmetry
How to submit a job
Along the navigation bar, hover over "Submit a design". You will see two options:
- via Nodes will bring you to an interactive board where you can manually position the helical rings using a 2D display either by drawing them (See Option A) or the circumferences and heights of each helix can be typed in (See Option B).
- via STL will allow you to upload a shape defined by an STL file (See Option C). DNAxiS will attempt to extract a 2D outline of the shape and automatically position rings to encapsulate the shape.
Option A: Draw Input
This interface currently supports structures with rotational symmetry only. Thus, geometries can be drawn in an unrevolved representation.
The left-hand side of the window can be used to place nodes. It is a cartesian plane where the horizontal axis corresponds to the circumference of the helical ring and the vertical axis corresponds to the height positioning of the ring. The right-hand side is a live preview of the shape after revolving drawn nodes. Only a half cross-section is displayed to better show the revolved geometry of each node.
Color labeling of nodes are a visual guide to ensure that adjacent helices are anti-parallel.
To change the direction of a helix, click on the node using the Right Mouse Button,
or in the text input interface, complement the direction bit (1 or 0).
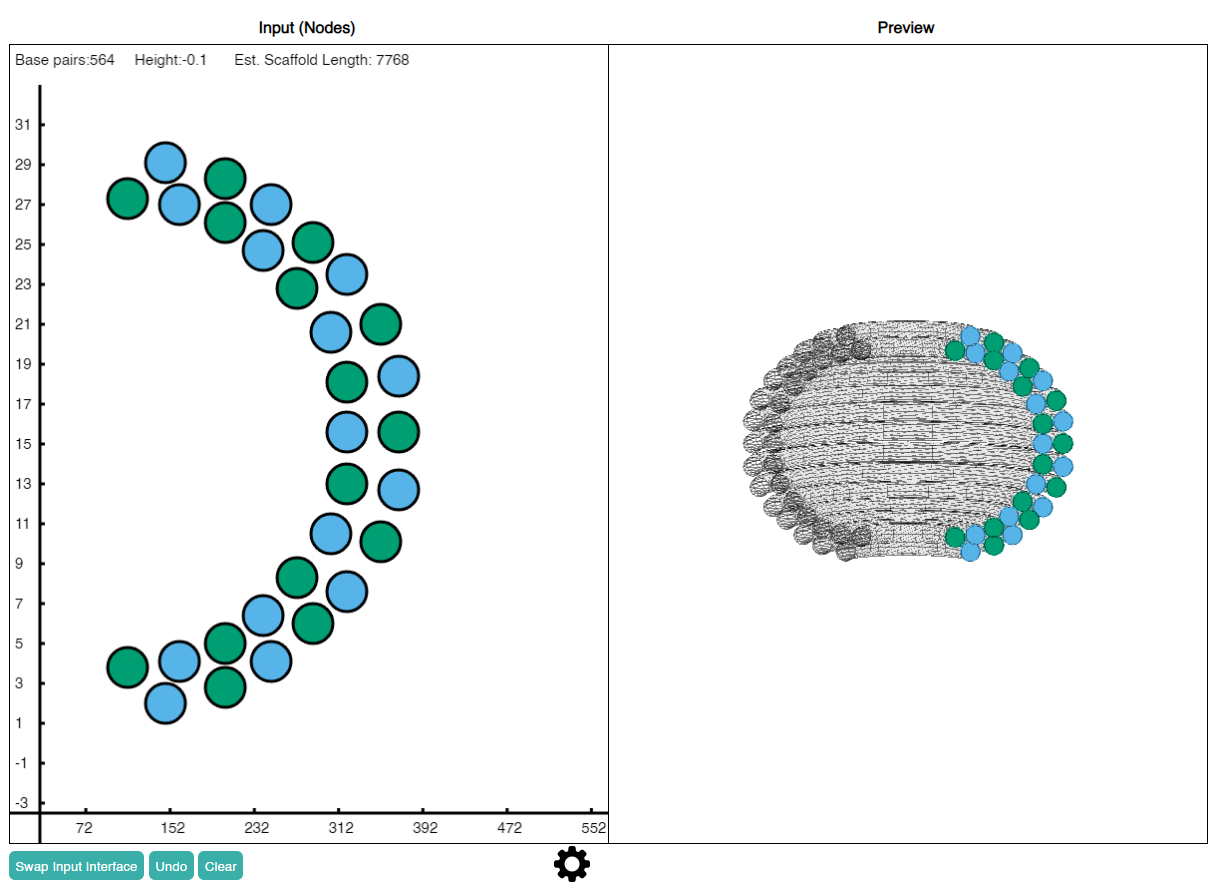 Draw nodes on the left. A 3D live preview in generated on the right.
Draw nodes on the left. A 3D live preview in generated on the right.
To fully draw a design:
- The first node can be placed anywhere on the board. Subsequent nodes must be placed to an existing node.
- For subsequent nodes, click on an existing node, which highlights the node in yellow.
- Another node will appear in white. This node will be placed in the direction of the cursor but is locked to an interhelical distance radius from the previous node and snaps to intervals of the crossover factor.
- Click again to place the node.
- Repeat this process to complete a design.
 New nodes will be draw in the direction of the cursor.
New nodes will be draw in the direction of the cursor.
Controls:
- Undo will erase the most recently placed node.
- Clear will erase the entire board.
- To adjust the crossover factor or interhlical distance snapping of new nodes, click on the Gears Icon at the bottom right of the board and select a different value. Note that changing this value will clear any existing work!
- Swap Input Interface will change to text input (Continue reading Option B).
- Mouse Wheel can be used to scroll the y-axes up or down.
Refer below to Submission Options to continue.
Option B: Text Input
The text input interface is accessible from the same screen as the Draw Input by clicking the Swap Input Interface button.
 The displays are equivalent.
The displays are equivalent.
This input is an alternate representation of drawing nodes with respect to the provided horizontal and vertical axis values. The format of each line is:
Circumference, Height, 5'-3' direction
Note: The 5'-3' direction of adjacent helices must be anti-parallel. 1 and 0 are opposite representations of the 5'-3' direction and should be alternated.
You may paste CSV data into the window to save and load structures. Snapping is not enforced for text input, except if you swap back to Draw Input.
Refer below to Submission Options to continue.
[Beta] Option C: By Stereolithography (STL) format
DNAxiS can attempt to automatically determine the profile you would like to revolve into an axially symmetric DNA origami nanostructure. This is best used when you want generated shapes to match existing shapes you are working with elsewhere.
- Design a structure in a 3D CAD design software of choice (e.g. SolidWorks, Blender, Autodesk Fusion 360, etc.)
- Export the object as STL.
- Import the structure by opening the STL file in DNAxiS.
- Use the preview window to check that DNAxiS is finding the correct outline of the structure.
- Click Send as input to automatically create a node design on the drawing board. This can be modified or submitted and following the same design pipeline as manual inputs.
If no changes are intended to be made, refer below to Submission Options to continue.
Submission Options
Several options are available that provide high-level control over the subsequent design process. The basic options are setting the desired staple length ranges and the preferred scaffold sequence. Both are set to common values by default.
A scaffold should be selected such that the structure produces the least length of unused scaffold. If more than the selected scaffold is required, the software will attempt to create a multi-scaffold structure using additional, orthogonal scaffold sequences. If the total structure is larger yet, the process will likely fail. Please submit feedback if you would like to use a longer, custom scaffold.
If you would like a link to download the design files sent to your email, you can optionally choose to provide your email address.
Advanced options are also available, but a set of defaults are provided that can typically produce structures with high reliability.
- Shape only will generate output without crossovers applied.
- Random scaffold sequence will override the application of a scaffold sequence and ignore lengths.
- Use greedy algorithm should be selected if “Use simulated annealing” is unchecked and will use a more basic crossover routing algorithm.
- Save steps will save the crossover configuration at each step. Only works for Simulated Annealing.
- Purge seams will attempt to remove crossovers on adjacent but separate pairs of helices.
- Ignore bounds and merge short strands will ignore the upper bound of staple lengths. This is useful if our implemented nicking algorithm is failing and a user can later manually add a single nick themselves.
- UV crosslinking will add thymines throughout the structure corresponding to UV crosslinking strategies as detailed by T. Gerling, M. Kube, B. Kick, H. Dietz, Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 4, eaau1157 (2018).
- Run simulated annealing will activate the simulated annealing algorithm for crossover routing. Do not use together with “Use greedy algorithm”.
- Log statistics will activate a verbose log file.
- Log validations increases running times to compile a report after running a series of integrity checks on the generated structure.
- Crossover alignment threshold controls the desired alignment during crossover routing.
- Same helix crossover spacing controls the permitted spacing during crossover routing when deciding between crossovers on the same edge.
- Adjacent helix crossover spacing controls the permitted spacing during crossover routing when deciding crossovers on adjacent edges.
- Crossover spacing lower bound alters the minimum accepted circumference in the geometry.
After setting options, click "Next" and proceed with Staple and Scaffold Routing.
Staple and Scaffold Routing
Each of the previous input options proceed into the same following pipeline.
Connections

The connections page asks you to declare all helices in the design that should be treated as adjacent. Staple crossovers will also be placed between the helixes as declared by the edges on this page.
An automatic routing is initially provided based on nearest neighbors seen from each helix that is within the interhelical distance. If this is incorrect, you may use the Clear button on the right to clear all edges (nodes will remain). The Undo button will clear the most recently drawn edge.
Click from one node to another to indicate that the algorithm should look for STAPLE crossovers between those two helices. You can also repeat this action to remove a specific edge. Hover over the "Help" button for a reminder on using this step of the interface.
Once you are finished, click "Next" and proceed to the Pathway step.
Pathway

The pathway page asks you to declare all helices in the design that should be routed in order by the scaffold strand. Adjacent helices as declared in the previous step are recorded and displayed as faint red lines as a reminder.
The node graph here cannot have a cycle. The tail of the scaffold strand is expected to be at the break. The graph must be otherwise fully connected. As before, you may use the Clear button on the right to clear all edges (nodes will remain). The Undo button will clear the most recently drawn edge.
Click from one node to another to indicate that the algorithm should look for SCAFFOLD crossovers between those two helices. You can also repeat this action to remove a specific edge.
The red guidelines indicate where you set staple crossovers in the previous step.
Scaffold crossovers should:
- Only be placed where there is a red line.
- Form a continuous path.
- Visit every node
Once you are finished, click "Next", which will submit the job to the server. Simple structures may take no longer than a minute, but larger and complex structures utilizing simulating annealing as the crossover routing algorithm can take up to an hour to complete. If you did not supply an email address, do not close the page. Once the processing is done, the page will be reloaded and provide a download link. The link is unique per job and cannot be recovered if you close the page.
The .conf and .top files that are downloaded as output can be viewed and also further edited in oxView.
Synthesizing your DNA Origami
To create your generated DNA origami,
- Purchase your chosen scaffold strand and the generated staple strands from vendors.
- Mix scaffolds and staples at a 1:10 ratio in the suggested conditions.
- Then anneal.
Below are several vendors you may consider and regions they serve as well as several example synthesis conditions from [1]:
Scaffolds:
Oligonucleotide synthesis for staples:
- Integrated DNA Technologies (Worldwide)
- Eurofins Genomics (India, Japan, and Europe)
- Sangon Biotech (China)
(Feel free to let us know your preferred vendors and they can also be added to the list.)
Sample protocols:
Disclaimer: There are not always consistent conditions for synthesizing your DNA origami, and it may be to your benefit to compare several at once.
Annealing buffer, 1XTAE-Mg2+
- 20 mM Tris base
- 10 mM acetic acid
- 0.5 mM ethylenediaminetetraacetic acid (EDTA)
- 12 - 20 mM Mg(OAc)2, pH 8.
Thermocycling programs
- 12 hour annealing protocol: Heat to 90°C for 5 min, jump to 86°C -5min and then decrease by 1°C/5min till 71°C, 70°C-15min and decrease by 1°C/15min to 40°C, 39°C-10min decrease by 1°C/10min to 26°C, 25°C-30min, jump to 20°C-15min, jump to 15°C-5min and jump to 10°C and maintained at that temperature.
- 24 hour annealing protocol: Heat to 90°C for 5 min, jump to 86°C-5min and then decrease by 1°C/5min till 76°C, 75°C-15min and decrease by 1°C/15min till 71°C, 70°C-20min decrease by 1°C/20min to 61°C, 60°C-30min decrease by 1°C/30min till 30°C, 29°C-20min decrease by 1°C/20min to 25°C, 24°C-15min decrease by 1°C/15min to 20°C, 19°C-10min decrease by 1°C/10min to 15°C, jump to 4°Cand pause maintained at that temperature.
- 37-hour annealing protocol: Heat to 80°C for 4 min and then decrease by 1°C/4min till 61°C, 60.5°C-30min and decrease by 0.5°C/30min till 34.5°C, 34°C-60min decrease by 1°C/60min to 24°C, and then pause maintained at that temperature.
- 48 hour annealing protocol: Heat to 90°C for 5 min, jump to 86°C-5min and then decrease by 1°C/5min till 81°C, 80°C-10min and decrease by 1°C/10min till 75°C, 74°C-30min decrease by 1°C/30min to 69°C, 68°C-40min decrease by 1°C/40min till 53°C, 52°C-60min decrease by 1°C/60min to 25°C, 24°C-80min decrease by 1°C/80min to 21°C, 20°C-30min,19°C/10min decrease by 1°C/10min to 15°C, jump to 4°Cand pause maintained at that temperature.
[1] Daniel Fu, Raghu Pradeep Narayanan, Abhay Prasad, Fei Zhang, Dewight Williams, John S. Schreck, Hao Yan, John Reif. Automated Design of 3D DNA Origami with Non-Rasterized 2D Curvature. (Under Revision), 2022.